Decoding the 2024 Nobel Prize in Chemistry: Predicting Protein Structures and Designing New Proteins with Computational Science
The 2024 Nobel Prize in Chemistry was awarded to Professor David Baker (University of Washington, USA), Sir Demis Hassabis (CEO and co-founder of Google DeepMind, UK), and Dr. John Michael Jumper (Director, Google DeepMind, UK) for their pioneering work in protein design and structure prediction.
Associate Professor Masahito Ohue from the School of Computing, a leading researcher in molecular design for drug discovery using computational science, reflects on the groundbreaking research that was awarded the Nobel Prize.
What is the novel research that won the 2024 Nobel Prize in Chemistry?
Ohue
This year’s Nobel Prize was awarded to three researchers for their extensive contributions to the field of protein design and structure prediction. Prof. Baker was recognized for his work in creating new proteins using computational methods, while Dr. Hassabis and Dr. Jumper received the prize for predicting protein structures from amino acid sequences.
Prof. Baker has significantly advanced both fields—protein design and structure prediction—Dr. Hassabis has also made important contributions to protein design. Their shared achievements in these two pivotal areas highlight the significance of their work. Moreover, it is particularly remarkable that tech companies like Google are now playing a key role in advancing life sciences research.

Why is predicting protein structures so significant?
Ohue
Most physiologic processes in the body are driven by proteins. Although proteins have diverse functions, they are all composed of 20 amino acids arranged in chains of varying lengths that fold into complex three-dimensional (3D) structures. For example, some influenza virus proteins have specific cavities that, when targeted by certain molecules, can inhibit the virus’s ability to multiply. This concept underpins the development of antiviral drugs like Tamiflu.
Molecular insights into protein structures are crucial for understanding their functions and developing new drugs. For decades, researchers have used experimental techniques such as X-ray crystallography and cryo-electron microscopy to determine protein structures. However, with the sheer number of proteins, studying them all experimentally is unfeasible. This challenge spurred the rise of computational methods in the 1990s, using algorithms and computers to predict protein structures efficiently.
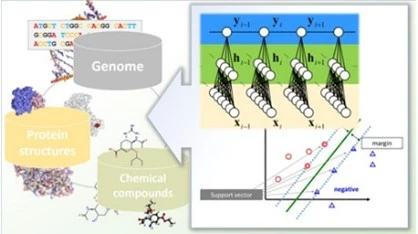
Can you elaborate on the contributions of Dr. Hassabis and Dr. Jumper?
Ohue
Since the 1990s, advancements in computing technology have accelerated research on protein structure prediction. However, early predictions were often inaccurate and ineffective. This changed in 2018 when Dr. Hassabis and Dr. Jumper developed the original version of AlphaFold. In November 2020, they released AlphaFold2, which achieved extraordinary results in the international CASP (Critical Assessment of Structure Prediction) competition, securing the top spot by a wide margin.
In July 2021, they published their findings on AlphaFold2 in Nature. This study demonstrated that AlphaFold2 utilized deep learning, a technique that had recently earned the Nobel Prize in Physics, through artificial neural networks. Remarkably, with a prediction accuracy exceeding 90%, AlphaFold2 holds the potential to uncover critical structural information and significantly accelerate drug discovery.
What are Prof. Baker’s contributions?
Ohue
While Prof. Baker has long been a pioneer in computational protein structure prediction, his Nobel recognition stems from his achievements in protein design. Allow me to walk you through his research step by step.
Proteins naturally adopt the most energetically stable 3D conformation, and proteins with similar amino acid sequences tend to have similar structures. Building on this principle, Prof. Baker created entirely new protein structures that are not found in nature. Rosetta was developed by Dr. Baker and his team, beginning in the late 1990s and culminating in the initial release of the software in 1998. Initially designed to predict protein structures, the software was later adapted to reverse the process, identifying amino acid sequences capable of forming stable 3D protein structure.
To validate their findings, Prof. Baker and his team synthesized the desired proteins in microorganisms using the identified amino acid sequences, confirming that the resulting structures perfectly matched their designs. One notable protein called Top7, consisted of 93 amino acids and had no natural analogs, marking a remarkable achievement. Published in 2003, this research captivated the scientific community. While the relationship between protein structure and function remains complex, this work paves the way for designing proteins with entirely new functions, potentially even creating new life forms.
What research are you currently conducting under the theme “Life Innovation through Computational Science”?
Ohue
You may have come across terms like antigens and antibodies in discussions about COVID-19. Antibodies are protective proteins produced by the immune system, while antigens are foreign proteins that signal the presence of external invaders in the body.
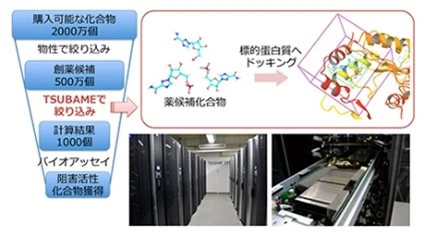
Antibody therapeutics can be designed to specifically bind to particular antigens, such as parts of the coronavirus, to neutralize them. In our lab, we utilize tools like Rosetta and AlphaFold2 to develop highly efficient antibody-based drugs that target specific antigens.
At Science Tokyo, we are fortunate to have access to the high-performance supercomputer TSUBAME 4.0, which enables large-scale computations. With many researchers specializing in protein structure prediction, we harness this collaborative environment to drive advances in medicine, chemistry, and life sciences through computational science.

* This article is based on the presentation given at Science Tokyo Nobel Prize Lecture, held online on Wednesday, November 20, 2024.
Discover More About Associate Professor Ohue's Work
Ongoing Similar Research at Science Tokyo
Computational Drug Discovery and Design Lab
Research aiming to understand biological phenomena using computational physical chemistry approaches, led by Professor Ryuichiro Ishitani, Associate Professor Yoshitaka Moriwaki, and their colleagues

Contact
Research Support Service Desk